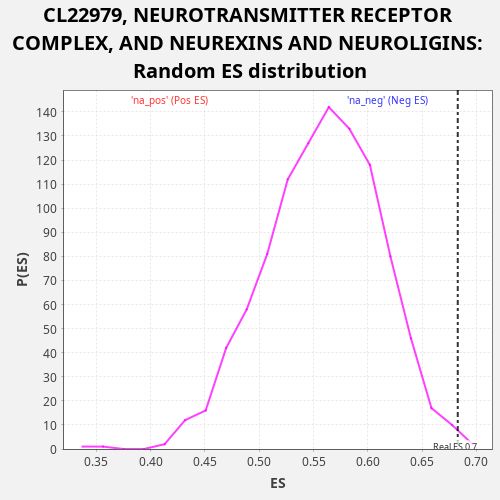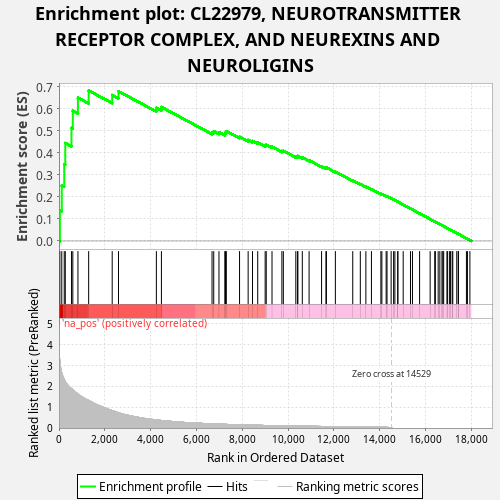
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CL22979, NEUROTRANSMITTER RECEPTOR COMPLEX, AND NEUREXINS AND NEUROLIGINS |
| Enrichment Score (ES) | 0.682902 |
| Normalized Enrichment Score (NES) | 1.2228599 |
| Nominal p-value | 0.004 |
| FDR q-value | 0.44353908 |
| FWER p-Value | 0.987 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | CACNG5 | 38 | 3.206 | 0.1400 | Yes |
| 2 | HOMER3 | 121 | 2.629 | 0.2519 | Yes |
| 3 | GRIN1 | 228 | 2.313 | 0.3485 | Yes |
| 4 | SLITRK5 | 274 | 2.224 | 0.4446 | Yes |
| 5 | GRIK4 | 544 | 1.886 | 0.5131 | Yes |
| 6 | CNTN4 | 603 | 1.828 | 0.5909 | Yes |
| 7 | NETO1 | 827 | 1.621 | 0.6503 | Yes |
| 8 | DLG2 | 1295 | 1.323 | 0.6829 | Yes |
| 9 | SYNGAP1 | 2322 | 0.831 | 0.6625 | No |
| 10 | HOMER2 | 2597 | 0.730 | 0.6796 | No |
| 11 | HTR3D | 4251 | 0.384 | 0.6044 | No |
| 12 | TJAP1 | 4471 | 0.359 | 0.6081 | No |
| 13 | GRIN2C | 6681 | 0.196 | 0.4936 | No |
| 14 | GRIK1 | 6755 | 0.193 | 0.4981 | No |
| 15 | CACNG8 | 6984 | 0.183 | 0.4935 | No |
| 16 | HTR3A | 7239 | 0.173 | 0.4870 | No |
| 17 | HTR3B | 7269 | 0.172 | 0.4930 | No |
| 18 | GRM8 | 7305 | 0.171 | 0.4986 | No |
| 19 | GRIN3A | 7882 | 0.152 | 0.4732 | No |
| 20 | GRIN3B | 8257 | 0.139 | 0.4585 | No |
| 21 | MDGA1 | 8448 | 0.133 | 0.4538 | No |
| 22 | FBXO40 | 8675 | 0.127 | 0.4468 | No |
| 23 | GRIA1 | 9000 | 0.119 | 0.4341 | No |
| 24 | GRIA4 | 9047 | 0.118 | 0.4367 | No |
| 25 | HTR3E | 9299 | 0.111 | 0.4277 | No |
| 26 | NETO2 | 9730 | 0.102 | 0.4082 | No |
| 27 | GRM1 | 9794 | 0.101 | 0.4092 | No |
| 28 | CBLN2 | 10339 | 0.091 | 0.3829 | No |
| 29 | CHRM5 | 10417 | 0.090 | 0.3825 | No |
| 30 | GRIN2D | 10426 | 0.090 | 0.3861 | No |
| 31 | GRM4 | 10622 | 0.086 | 0.3790 | No |
| 32 | CLSTN3 | 10918 | 0.081 | 0.3661 | No |
| 33 | CHRM3 | 11463 | 0.072 | 0.3390 | No |
| 34 | NLGN2 | 11654 | 0.069 | 0.3314 | No |
| 35 | SHISA6 | 11674 | 0.069 | 0.3334 | No |
| 36 | NRXN2 | 12065 | 0.063 | 0.3145 | No |
| 37 | CDH9 | 12824 | 0.053 | 0.2745 | No |
| 38 | CNIH3 | 13154 | 0.048 | 0.2583 | No |
| 39 | OLFM2 | 13397 | 0.045 | 0.2468 | No |
| 40 | GRIK2 | 13640 | 0.042 | 0.2352 | No |
| 41 | GRIK3 | 14052 | 0.035 | 0.2138 | No |
| 42 | SHANK1 | 14095 | 0.034 | 0.2130 | No |
| 43 | NLGN4X | 14282 | 0.029 | 0.2039 | No |
| 44 | HOMER1 | 14320 | 0.028 | 0.2031 | No |
| 45 | LHFPL4 | 14500 | 0.016 | 0.1938 | No |
| 46 | NRXN3 | 14607 | -0.000 | 0.1879 | No |
| 47 | CBLN4 | 14663 | -0.000 | 0.1848 | No |
| 48 | DLGAP4 | 14788 | -0.000 | 0.1779 | No |
| 49 | DLG3 | 14794 | -0.000 | 0.1776 | No |
| 50 | CBLN1 | 15026 | -0.000 | 0.1647 | No |
| 51 | DLGAP3 | 15344 | -0.000 | 0.1471 | No |
| 52 | GRIA2 | 15431 | -0.000 | 0.1423 | No |
| 53 | DLG4 | 15742 | -0.000 | 0.1250 | No |
| 54 | GRID2 | 16203 | -0.000 | 0.0993 | No |
| 55 | SHANK2 | 16404 | -0.000 | 0.0882 | No |
| 56 | LRRTM1 | 16430 | -0.000 | 0.0868 | No |
| 57 | PTCHD1 | 16553 | -0.000 | 0.0800 | No |
| 58 | CACNG2 | 16626 | -0.000 | 0.0760 | No |
| 59 | GRM5 | 16718 | -0.000 | 0.0709 | No |
| 60 | NLGN1 | 16759 | -0.000 | 0.0687 | No |
| 61 | DLGAP1 | 16793 | -0.000 | 0.0668 | No |
| 62 | VWC2L | 16939 | -0.000 | 0.0587 | No |
| 63 | CNIH2 | 16955 | -0.000 | 0.0579 | No |
| 64 | HTR3C | 17042 | -0.000 | 0.0531 | No |
| 65 | NRXN1 | 17089 | -0.000 | 0.0505 | No |
| 66 | GRID1 | 17164 | -0.000 | 0.0464 | No |
| 67 | GRIN2A | 17191 | -0.000 | 0.0450 | No |
| 68 | SHISA7 | 17365 | -0.000 | 0.0353 | No |
| 69 | GRM7 | 17434 | -0.000 | 0.0315 | No |
| 70 | NLGN3 | 17435 | -0.000 | 0.0315 | No |
| 71 | SHISA9 | 17787 | -0.000 | 0.0119 | No |
| 72 | SHANK3 | 17830 | -0.000 | 0.0096 | No |
| 73 | GRIN2B | 17938 | -0.000 | 0.0036 | No |